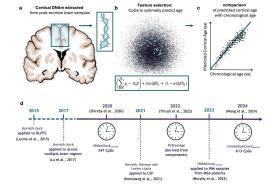
Dr Conole’s work sits at the intersection of artificial intelligence, genetics, and neuroscience, demonstrating how AI-driven approaches can unlock new insights into the biology of ageing and disease.
The paper, Epigenetic clocks and DNA methylation biomarkers of brain health and disease, looks at how chemical markers on our DNA - shaped by both genetics and lifestyle - can be used to track the pace of ageing in the brain.
Epigenetics describes how chemical modifications to our DNA regulate gene activity without altering the genetic code itself. These modifications are dynamic and responsive to the way we live: diet, exercise, stress and alcohol consumption all leave traces at the molecular level, influencing how we age. Over time, epigenetics captures the biological imprint of our lifestyles, embedding it into our cells in ways that affect disease risk and long-term health.
One key process is DNA methylation, where small chemical tags attach to DNA at specific sites in the genome. These methylation patterns shift in predictable ways over a lifetime, making them powerful markers of biological ageing. By analysing DNA methylation with machine learning, researchers can create “epigenetic clocks” that estimate a person’s biological age. Unlike chronological age, which simply counts years, biological age reflects how quickly or slowly the body is actually ageing. A faster “ticking” biological clock has been linked to higher risks of conditions such as heart disease, cancer and dementia.
Most epigenetic clocks so far have been trained on blood samples, as these are easier to collect. However, the new review highlights the importance of developing brain-specific clocks, trained directly on DNA from brain tissue. These brain clocks capture molecular changes linked to ageing more precisely and show stronger associations with neurodegeneration than blood-based measures.
Although still at the research stage, brain-based clocks offer huge promise. They could one day help clinicians detect early deviations from healthy cognitive ageing, monitor brain health more accurately, and guide strategies to preserve memory and thinking skills later in life.
The full article is available in Nature Reviews Neurology: Epigenetic clocks and DNA methylation biomarkers of brain health and disease.